From Data to Diagnosis: How IBM LinuxONE Powered Our Datathon Winning AKI Dialysis Prediction Project
Authors: Harshavardhan MG (Team leader) - BE in AIML, Global Academy of Technology, harshavardhangowda2004@gmail.com, P Praveen Raj - BE in AIML, Global Academy of Technology, 7220praveen@gmail.com, Rishith P - BE in AIML, Global Academy of Technology, rishithprasanna264@gmail.com, Suhas S Gowda - BE in AIML, Global Academy of Technology , Jeevan K Thejas - BE in AIML, Global Academy of Technology
Mentors: Vineet Dumir - Senior Solution Architect, AI on IBM Z, IBM Labs, Bangalore, INDIA, Vineet.Dumir@ibm.com, Vineet Dumir Sudharsana Srinivasan - Senior Technical Program Manager, Pleasanton, CA, Sudharsana.Srinivasan@ibm.com, Sudharsana Srinivasan
Introduction
Hello everyone! We're Team Alpha, and we’re excited to share our journey in developing an innovative AI/ML solution for predicting early dialysis needs in ICU patients with Acute Kidney Injury (AKI). This project represents a critical intersection of advanced machine learning techniques and real-time clinical decision-making in intensive care settings.
To prepare for the datathon, IBM mentors conducted workshops introducing AI/ML frameworks on IBM Z. We gained hands-on experience with IBM LinuxONE Community Cloud (L1CC), setting up machine learning environments using Docker containers on Ubuntu Linux. With easy-to-follow tutorials, we quickly deployed popular AI frameworks like TensorFlow and PyTorch. It was exciting to see how seamlessly familiar AI tools could run on IBM Z’s s390x architecture, reinforcing its adaptability for enterprise AI workloads.
In this blog, we’ll take you through our journey of developing and deploying an AI/ML solution on IBM Z systems—from the initial idea to a fully operational model.
The Problem
Predicting early dialysis needs for ICU patients with Acute Kidney Injury (AKI) is a crucial challenge in intensive care settings where timing is everything.
Why is this important? When patients develop AKI in the ICU, medical teams face a crucial dilemma:
Currently, this decision relies heavily on manual monitoring and individual judgment, making it prone to delays and variations in care. The challenge is compounded by the sheer volume of patient data—from vital signs to laboratory results—that must be analyzed in real time.
Our goal was to develop an AI-powered solution capable of processing this complex data and providing timely, accurate predictions to support medical decision-making. This isn’t just about building a model; it’s about potentially saving lives through better-timed interventions and more efficient resource allocation in critical care settings.
The Dataset
Our project leverages a comprehensive ICU patient dataset curated specifically for AKI prediction. The dataset includes key ICU patient metrics such as heart rate, blood pressure, creatinine levels, AKI stage, anion gap, and PT max—all critical indicators for early dialysis prediction.
Given that patients requiring early dialysis intervention are fewer in number, we used specialized sampling techniques to address class imbalance:
-
SMOTE (Synthetic Minority Over-sampling Technique) – Generates synthetic samples for the minority class.
-
Class Weights – Assigns higher penalties for misclassifying the minority class (class_weight='balanced').
Data Preprocessing Implementation
# Step 1: Data Preprocessing Pipeline
preprocessor = ColumnTransformer([
('num', StandardScaler(), numeric_columns),
('cat', OneHotEncoder(drop='first', sparse_output=False), categorical_columns)
])
# Step 2: Feature Selection
feature_selector = SelectFromModel(
RandomForestClassifier(n_estimators=100, random_state=42),
max_features=20
)
# Step 3: Complete Pipeline with SMOTE and Class Weights
pipeline = Pipeline([
('preprocessor', preprocessor),
('feature_selector', feature_selector),
('smote', SMOTE(random_state=42)),
('classifier', LogisticRegression(random_state=42, class_weight='balanced'))
])
Model Implementation
To predict early dialysis needs in ICU patients, we implemented a suite of machine learning models, including:
-
Logistic Regression – Offers interpretable results with cross-validated hyperparameter tuning.
-
Random Forest – Captures complex feature interactions and identifies key AKI predictors.
-
XGBoost – A powerful gradient boosting technique that enhances performance through grid search optimization.
-
Ensemble Techniques – Combines multiple models using a voting classifier for enhanced prediction accuracy.
Each model underwent rigorous evaluation using metrics such as accuracy, precision, recall, F1-score, and ROC AUC. Our best-performing model, XGBoost, achieved an F1-score of 0.87 and an AUC of 0.92, ensuring reliable dialysis predictions in ICU settings.
The table below presents a comparative analysis of different machine learning models based on their classification performance metrics.
| Model Application |
Accuracy |
Class 0
Precision
|
Class 0
Recall
|
Class 0
F1
|
Class 1
Precision
|
Class 1
Recall
|
Class 1
F1
|
| XGBoost Application |
78% |
0.39 |
0.43 |
0.41 |
0.87 |
0.85 |
0.86 |
| Random Forest Application |
~75% |
0.29 |
0.29 |
0.29 |
0.85 |
0.85 |
0.85 |
| Logistic Regression Application |
63% |
0.26 |
0.57 |
0.35 |
0.87 |
0.64 |
0.74 |
| Ensemble (Voting) |
80% |
0.36 |
0.14 |
0.20 |
0.84 |
0.95 |
0.89 |
Workflow of our Project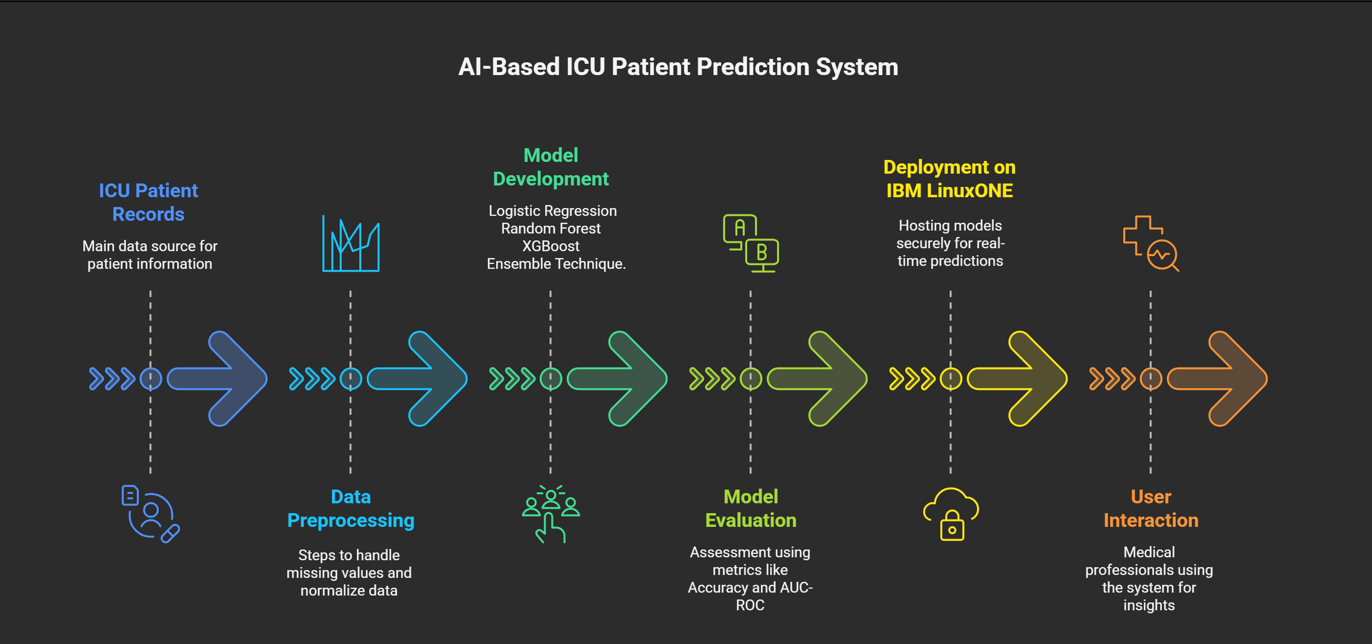
Deploying to IBM LinuxONE Community Cloud (L1CC)
The deployment phase was a critical milestone, marking the transition from model development to real-world application. The IBM Z platform proved to be the ideal environment for operationalizing our AKI prediction models.
Thanks to IBM Z’s compatibility with open standards, we seamlessly transferred our trained models (Logistic Regression, Random Forest, XGBoost, and ensemble models) to the L1CC environment.
Key benefits of IBM Z deployment:
-
High-speed inference: Model predictions were generated in under 1 second—significantly faster than local execution.
-
Scalability & reliability: The enterprise-grade infrastructure ensured consistent model performance.
-
Enhanced security: IBM Z’s built-in encryption provided a secure environment for handling sensitive medical data.
The successful deployment on IBM LinuxONE Community Cloud demonstrated the real-world feasibility of our system for clinical applications, where timely predictions can make a life-saving difference.
ROC-AUC Graph

Here is the video blog of our incredible journey with IBM and video description of our project.
Conclusion
Our journey in developing the Early Dialysis Prediction System has been both challenging and rewarding. By implementing a combination of Logistic Regression, Random Forest, XGBoost, and ensemble techniques, we successfully created a robust AI system for predicting dialysis needs in ICU patients with AKI.
Implementation Representation

Key Takeaways:
-
Handles complex medical data preprocessing with advanced techniques like SMOTE & feature selection.
-
Achieves high predictive accuracy using multiple ML models with XGBoost leading in performance.
-
Deploys seamlessly on IBM Z, ensuring fast inference, enterprise scalability, and security.
Looking ahead, we plan to:
-
Expand our dataset to improve generalization.
-
Enhance model interpretability to support better clinical adoption.
-
Validate our solution in real-world clinical settings to assess practical impact.
For now, we’re proud to have developed an AI-powered system that could assist medical professionals in making timely dialysis decisions. A huge thank you to our mentor Vineet Dumir and everyone who supported this project!
Until next time,
Team Alpha